Section: New Results
Multiscale Visualization and Scale-Adaptive Modification of DNA Nanostructures
Participants : Haichao Miao [TU Wien, Austria, and Austrian Institute of Technology, Vienna, Austria] , Elisa de Llano [Austrian Institute of Technology, Vienna, Austria] , Johannes Sorger [Complexity Science Hub Vienna, Austria] , Yasaman Ahmadi [Austrian Institute of Technology, Vienna, Austria] , Tadija Kekic [Austrian Institute of Technology, Vienna, Austria] , Tobias Isenberg [correspondant] , M. Eduard Gröller [TU Wien, Austria] , Ivan Barišic [Austrian Institute of Technology, Vienna, Austria] , Ivan Viola [TU Wien, Austria and KAUST, Kingdom of Saudi Arabia] .
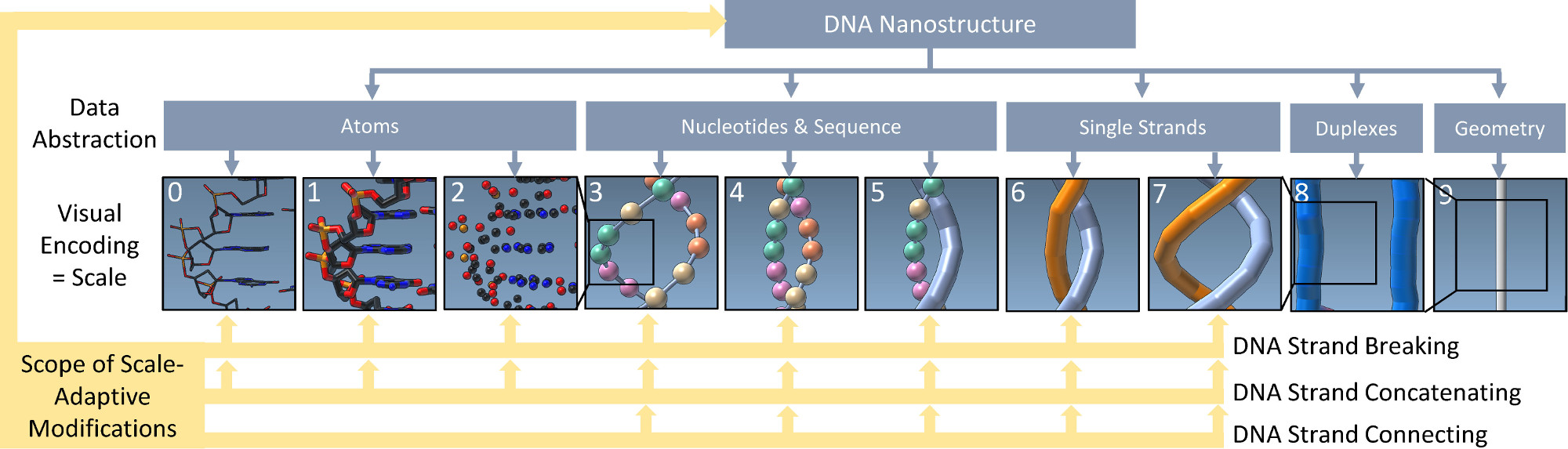
We presented an approach to represent DNA nanostructures in varying forms of semantic abstraction, describe ways to smoothly transition between them, and thus create a continuous multiscale visualization and interaction space for applications in DNA nanotechnology. This new way of observing, interacting with, and creating DNA nanostructures enables domain experts to approach their work in any of the semantic abstraction levels, supporting both low-level manipulations and high-level visualization and modifications. Our approach allows them to deal with the increasingly complex DNA objects that they are designing, to improve their features, and to add novel functions in a way that no existing single-scale approach offers today. For this purpose we collaborated with DNA nanotechnology experts to design a set of ten semantic scales (see Figure 8). These scales take the DNA's chemical and structural behavior into account and depict it from atoms to the targeted architecture with increasing levels of abstraction. To create coherence between the discrete scales, we seamlessly transition between them in a well-defined manner. We used special encodings to allow experts to estimate the nanoscale object's stability. We also added scale-adaptive interactions that facilitate the intuitive modification of complex structures at multiple scales. We demonstrate the applicability of our approach on an experimental use case. Moreover, feedback from our collaborating domain experts confirmed an increased time efficiency and certainty for analysis and modification tasks on complex DNA structures. Our method thus offers exciting new opportunities with promising applications in medicine and biotechnology.
More on the project Web page: https://tobias.isenberg.cc/VideosAndDemos/Miao2018MVS.